2023-08-23 Setting up CRISPResso2 as an AWS Batch Job: A Comprehensive Guide
Setting up CRISPResso2 as an AWS Batch job offers a scalable and efficient solution for processing CRISPR-generated sequencing data
Overview
CRISPR technology has revolutionized molecular biology and genetic engineering, allowing scientists to edit DNA with precision. As the field has advanced, so have the tools used to analyze and interpret CRISPR-generated data. CRISPResso2 is one such tool, and in this blog post, we'll guide you through setting it up as an AWS Batch job. But first, let's begin with some background information on CRISPR and CRISPResso2.

Introduction to CRISPR
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a revolutionary gene-editing technology that allows precise modification of DNA in living organisms. This technology, derived from the immune system of bacteria and archaea, has opened up countless possibilities in genetic research, biotechnology, and medicine. CRISPR systems are composed of two main components: a guide RNA (gRNA) and a nuclease enzyme (usually Cas9). The gRNA guides the nuclease to the target DNA sequence, where it induces a cut, leading to gene modifications.
Introduction to CRISPResso2
CRISPResso2 is a powerful computational tool developed by the Pinello lab at Boston University that facilitates the analysis of CRISPR-Cas9 generated sequencing data. It allows researchers to assess the efficiency and accuracy of genome editing events, detect and quantify indels (insertions and deletions), and visualize the outcomes of CRISPR experiments. CRISPResso2 has a comprehensive set of features, making it an essential tool for scientists working in the field of genome editing.
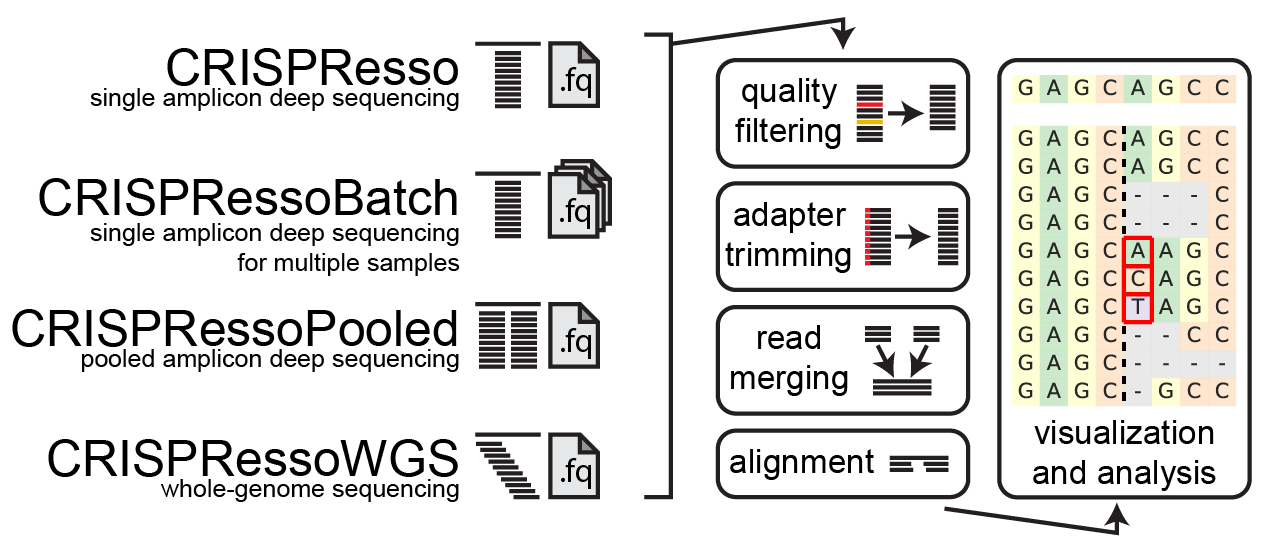
Challenges with Installing CRISPResso2 and Computational Intensity
Before we delve into setting up CRISPResso2 on AWS Batch, it's essential to highlight the challenges associated with installing CRISPResso2 on a local machine. CRISPResso2 requires a complex set of dependencies, and managing these dependencies, especially with Conda, can be challenging and time-consuming. Additionally, analyzing CRISPR data can be computationally intensive, which may strain the resources of a regular desktop or server. To address these challenges, it's advantageous to use cloud computing resources.
Using the Docker Image pinellolab/crispresso2
pinellolab/crispresso2An elegant solution to avoid the complexities of installing CRISPResso2 and manage computational intensity is to use the Docker image pinellolab/crispresso2. Docker allows you to package an application and its dependencies into a container, ensuring consistent execution across different environments. By utilizing this pre-built Docker image, you can create your CRISPResso2 pipeline without the need for manual installations.
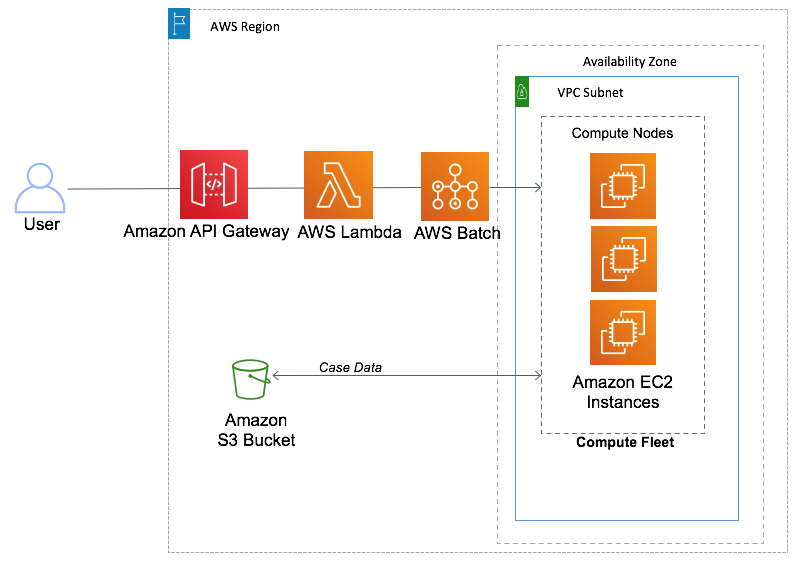
Here are the basic steps to set up CRISPResso2 as an AWS Batch job using the Docker image:
Step 1: Set Up AWS Batch Environment
Create an AWS Batch job queue, compute environment, and job definition as per your requirements.
Step 2: Create a Docker Container
Write a Dockerfile or use an existing one to create a Docker container that includes the pinellolab/crispresso2 image and any additional dependencies your analysis may require.
Step 3: Define Your Batch Job Script
Create a batch job script that specifies the analysis parameters, input data location, and output data destination.
Step 4: Submit the AWS Batch Job
Submit your job to AWS Batch, specifying the Docker container and script to execute.
Using this approach, you can easily scale your CRISPResso2 analysis by leveraging the computational resources available on AWS Batch without worrying about the installation intricacies.
Integration with Labii for Seamless Workflow
For researchers and labs looking to streamline their CRISPR data analysis, consider integrating CRISPResso2 as an AWS Batch job with Labii, a cloud-based laboratory information management system (LIMS). Labii provides a user-friendly interface for managing experiments, data, and results.
By integrating CRISPResso2 as an AWS Batch job with Labii, you can:
Manage Data: Easily upload and organize your sequencing data within Labii.
Configure Analyses: Define analysis parameters and settings directly within Labii.
Automate Execution: Initiate CRISPResso2 jobs on AWS Batch from the Labii interface, reducing manual effort.
Access Results: Retrieve and visualize CRISPResso2 results seamlessly within Labii.
This integration enhances collaboration, simplifies data management, and accelerates the pace of research by combining the power of CRISPResso2 with the convenience of Labii.
Conclusion
In conclusion, setting up CRISPResso2 as an AWS Batch job offers a scalable and efficient solution for processing CRISPR-generated sequencing data. By utilizing Docker containers and exploring integration options like Labii, you can streamline your CRISPR data analysis workflow and focus on advancing your research.
To learn more, schedule a meeting with Labii representatives (https://call.skd.labii.com) or create an account (https://www.labii.com/signup/) to try it out yourself.